5 minute read.Preprints growth rate ten times higher than journal articles
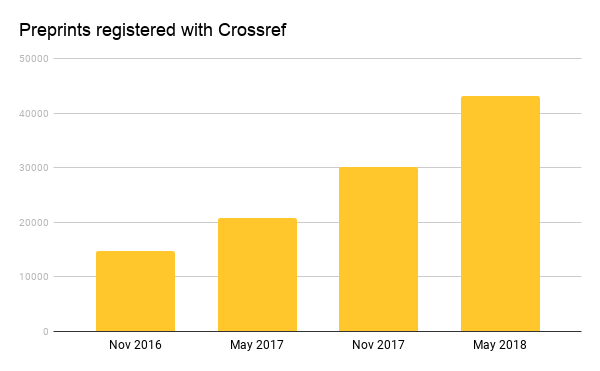
The Crossref graph of the research enterprise is growing at an impressive rate of 2.5 million records a month - scholarly communications of all stripes and sizes. Preprints are one of the fastest growing types of content. While preprints may not be new, the growth may well be: ~30% for the past 2 years (compared to article growth of 2-3% for the same period). We began supporting preprints in November 2016 at the behest of our members. When members register them, we ensure that: links to these publications persist over time; they are connected to the full history of the shared research results; and the citation record is clear and up-to-date.
Summary
As of May 24, 2018 we have 44,388 works (see API query https://0-api-crossref-org.libus.csd.mu.edu/types/posted-content/works with a json viewer) registered as posted content. Today that number is over 150k. Preprints are part of this record type category, which is meant to house scholarly outputs that have been posted online and intended for publication in the future.
For a more granular view, see the monthly stats captured by Jordan Anaya in PrePubMed. This data is based on a slightly different set of preprint repositories, though both show the same trends.
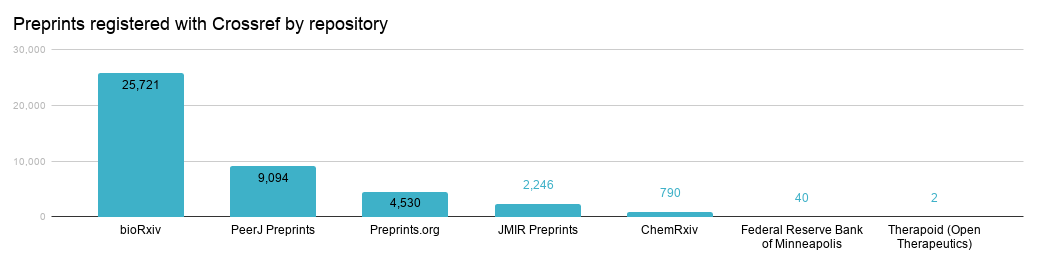
The figure below shows the preprints registered with Crossref, broken down by repository.
We eagerly await our newest preprints member, Center for Open Science, who will soon be registering the preprints from their 18 community archives with us (~9k preprints total to date).
We accept a range of metadata for the preprints registered with us, including:
- Repository name & hosting platform
- Contributor names & ORCID iDs
- Title
- Dates (posted, accepted)
- License
- Funding
- Abstract
- Relations
- References
As with all resource/record types, certain metadata is required, though others are optional. We encourage full coverage of metadata in the record where applicable and possible. So what are publishers including in their posted content records? The summary view is as follows:
- License: 9926 (json), 22% (PeerJ Preprints, ChemRxiv)
- Funder: 0 (json), 0%
- ORCID: 19309 (json), 44% (bioRxiv, PeerJ Preprints, Preprints.org, ChemRxiv)
- Abstracts: 35874 (json), 81% (bioRxiv, PeerJ Preprints, ChemRxiv)
- References: 1921 (json):, 4% (JMIR)
Compared to all the published content registered with us over time, preprints have above average coverage of ORCID iDs deposited and show well above average with abstract metadata. However, they are significantly lagging behind with depositing references, license, and funding metadata. (See a summary of the full corpus stats taken two months ago in the blog post, A Lustrum over the Weekend.
Preprint-article pairs
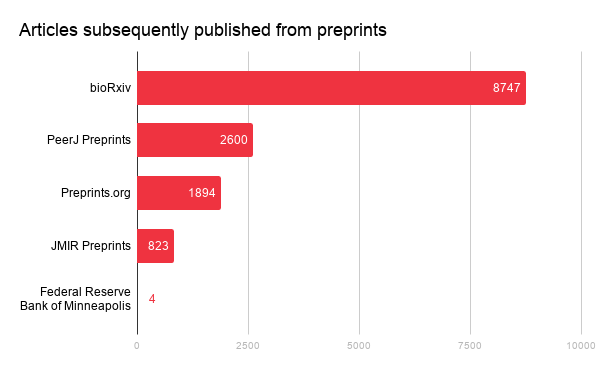
Members registering preprints have an obligation to update the metadata record when a journal article is subsequently published, to clearly identify this work. This pairing is passed on to our metadata users: indexing platforms; recommendations engines; platforms; tools, etc. which pull from our APIs. (The preprint landing page also must link to the article.) As such, the preprint-article pairings are amassing as each week passes. We currently have a total of 12983 (json) preprints connected to articles. The figure below provides the counts based on repository.
Citations
We can see from preprint Cited-by counts that researchers are indeed citing preprints in their articles. This practice is an extension of the common citation behavior to provide evidence for and credit to previous work, a natural consequence of work shared with their peers. The most highly cited preprint papers (json) as of May 24, 2018 are as follows. In some cases, a subsequent paper was published from the results shared in the preprint. These have also accrued citations in their own right and these are also indicated in the table below.
| No. | Cited-by | Preprint DOI | Preprint title | Date | Subsequent journal article | Citations of journal article |
|---|
| 1 | Cited-by 72 | https://0-doi-org.libus.csd.mu.edu/10.1101/005165 | qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots | May 14, 2014. | n/a | n/a |
| 2 | Cited-by 63 | https://0-doi-org.libus.csd.mu.edu/10.1101/002824 | HTSeq - A Python framework to work with high-throughput sequencing data | August 19, 2014 | Bioinformatics, https://0-doi-org.libus.csd.mu.edu/10.1093/bioinformatics/btu638 | 2372 |
| 3 | Cited-by 43 | https://0-doi-org.libus.csd.mu.edu/10.1101/030338 | Analysis of protein-coding genetic variation in 60,706 humans | May 10, 2016 | Nature, https://0-doi-org.libus.csd.mu.edu/10.1038/nature19057 | 1598 |
| 4 | Cited-by 38 | https://0-doi-org.libus.csd.mu.edu/10.1101/002832 | Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 | November 17, 2014 | Genome Biology, https://0-doi-org.libus.csd.mu.edu/10.1186/s13059-014-0550-8 | 3284 |
| 5 | Cited-by 32 | https://0-doi-org.libus.csd.mu.edu/10.1101/021592 | Salmon provides accurate, fast, and bias-aware transcript expression estimates using dual-phase inference | August 30, 2016 | Nature Methods, https://0-doi-org.libus.csd.mu.edu/10.1038/nmeth.4197 | 112 |
| 6 | Cited-by 22 | https://0-doi-org.libus.csd.mu.edu/10.1101/012401 | DensiTree 2: Seeing Trees Through the Forest | December 8, 2014 | n/a | n/a |
| 7 | Cited-by 21 | https://0-doi-org.libus.csd.mu.edu/10.1101/011650 | FusionCatcher - a tool for finding somatic fusion genes in paired-end RNA-sequencing data | November 19, 2014 | n/a | n/a |
| 8 | Cited-by 19 | https://0-doi-org.libus.csd.mu.edu/10.1101/048991 | Analysis of shared heritability in common disorders of the brain | September 6, 2017 | n/a | n/a |
| 9 | Cited-by 18 | https://0-doi-org.libus.csd.mu.edu/10.1101/006395 | Error correction and assembly complexity of single molecule sequencing reads | June 18, 2014 | n/a | n/a |
| 10 | Cited-by 18 | https://0-doi-org.libus.csd.mu.edu/10.1101/032839 | Spread of the pandemic Zika virus lineage is associated with NS1 codon usage adaptation in humans | November 25, 2015 | n/a | n/a |
The relationship between preprints and the proceeding publication is an interesting area that is not yet well understood. We invite the community to analyze the Crossref metadata using the REST API in concert with other datasets. For example, the citation lifecycle for these two research products has been one of speculation so far without a systematic investigation into patterns and timeframes of preprint citations and those of its succeeding article across the corpus. Here, submission dates would be critical data to this research question as publication windows vary significantly by publisher and by paper.